Release version 0.b1
Summary description
The test procedure comprises three steps, i.e. (i) saliva collection and filtration, (ii) nucleic acid extraction and amplification, and (iii) detection. As for the amplification, we opted for a MNAzyme (Multicomponent DNAzyme) catalyzed amplification, due to the non proteinaceous nature, the large temperature range of the catalytic activity, the low cost and prompt availability for large quantities, which are all necessary characteristics to meet the requirements. The MNAzyme catalyzed reaction results in the production of a DNA fragment that, in the presence of hemin, assembles into a G-quadruplex structure with peroxydase activity (known as HRP-mimicking). Hence, the amplification product is loaded in a plastic (polystyrene) microfluidic device where it is purified through paper chromatografy and further incubated to associate with hemin. Then the peroxydase activity is exploited in a plasmonic detection step. To this end, a solution containing gold(III) is loaded in the reagent reservoirs and distributed in the test well to promote the formation of gold nanoparticles (AuNP). Finally, the device is pictured with a smartphone and the results sent for remote analysis.
1. Saliva is chosen as the sampling material for the ease in self sampling and on the basis of existing records on SARS-CoV and SARS-CoV-2 (Pfaffe et al., 2011; Arunachalam et al., 2019; Low et al., 2020; To et al., 2020; Wang et al., 2020). A discussion page to evaluate the opportunity of choosing different sampling material is here
2. After collection in a Polyethylene vial the sample is filtered on a .45 μm membrane by squeezing the vial and then mixed with the lysis/amplification buffer B1. The filtration is carried out to select virions and discard cells, allowing a simplified nucleic acid extraction. To this end, the Proteinase K - SDS method was chosen because there is no protein involved in the following amplification step, hence the lysis and signal amplification can be performed together. A discussion page to evaluate the opportunity of choosing a different method for the preparation of nucleic acid is here
3. An MNAzyme (Multicomponent Nucleic Acid enzyme; Mokany et al., 2010) amplification is carried out with a modification of the method of Xie et al. (2020). The reaction is kept at 37°C for 120 minutes; the suggested methods to perform the incubation at 37°C in household conditions are yet to be determined.
Note: Further modifications for adapting the system to room temperature and increase the catalytic performance as done for the 10-23 DNAzyme (Ven et al., 2019; Safdar et al., 2020) would be highly desirable. A reduction in the incubation time would be highly desirable. A discussion page on those topics and /or to evaluate the opportunity of choosing different molecular signal processing methods is here
The MNAzyme amplification uses as target an RNA sequence region within gene N (positions 28.274 to 29.533) as it is conserved in all SARS-CoV-2 sequences available from NCBI at the date of this report, March 24, 2020. The N gene of SARS-CoV-2 is about 10 times more sensitive in detecting positive clinical specimens than the ORF-1b gene since it is encoded by subgenomic mRNA (Chu et al., 2020). A discussion page to evaluate the opportunity of choosing different target is here
The MNAzyme amplification results in the accumulation of copies of a 27mer DNA oligo containing the sequence of the HRP-mimiking DNAzyme (Xie et al., 2020) with the adenine base extended at the 3′end to enhance its peroxidase activity (Li et al., 2016; Chen et al., 2019).
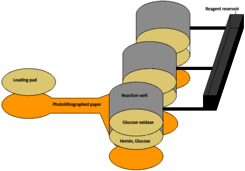
4. Following amplification the resulting DNA is purified by paper cromatography, that was printed using the well assessed UV lithography with SU-8 photoresist on Whatman cellulose #1 (Martinez et al., 2007). The small DNAzyme migrates fast and is readily available at the elution zone (Fronczek et al., 2014).
Note: Length of the channel and volume to load must be optimized.
5. At the end of the chromatography run the solvent rehydrates hemin, glucose and glucose oxidase that were entrapped in cellulose disks at the elution zones. There are three elution zones with cellulose disks with slightly different content, corresponding to Test, Negative Control, and Positive Control zones (see Material section). Upon rehyidration, the glucose oxydase starts producing hydrogen peroxide while the 27mer resulting from the amplification assembles with hemin into a G-quadruplex with peroxidase activity (HRP-mimiking DNAzyme). The H2O2 production is allowed to proceed for 15 minutes then, upon the addition of 50 µl per well of gold(III) cloride solution in MES, AuNP are produced according to de la Rica & Stevens (2012, 2013).
6. The color developped in the reaction wells is photographed four times within 15 minutes and the photographs sent for remote processing by a dedicated smartphone App. The image is processed according to Soda and Bakker (2019).
Here is how the diagnostic test will be carried out by the final user:
Do not eat or drink for at least 30 minutes before testing.
1. Start the App on your smartphone.
2. Push the cap of vial 1 to break the inner membrane and mix vigourosly untill the powder is dissolved
3. Prepare the saliva collector: open vial2 and insert the funnel shaped collector
4. Collect at least 0.5 ml saliva into vial2. Dismount the funnel and mount the filter-equipped dropper.
5. Open vial1, then squeeze vial2 to drop 0.2 ml into vial1.
6. Close vial1 and allow it for 120 minutes at 37 °C (keep in hands or close to body with a bandage). Press the green button on the App.
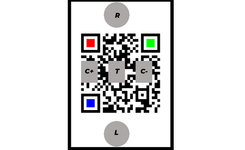
7. Open vial1 and mount the clean dropper, then drop 0.1 ml (two drops) into the loading well (marked L) of the test box (only 2 drops; take care to avoid overloading). Press the Yellow button on the App.
8. Wait for 15 minutes
9. Turn the box, peel off the paper strip, insert the box in the holder and press untlil the box is firmly locked.
10. Push the cap of vial3 to break the inner membrane and mix vigourosly untill the powder is dissolved. Pour the entire content of vial3 into the reagent well (marked R) of the test box. Press the red button on the App.
11. When requested by the app, picture the QR code on the top of the box four times in 15 min. Wait for the result to appear on the screen.
Plastic vial with dehydrated B1 buffer (vial 1), cap with filter and dropper (for vial1), Plastic vial with dropper (vial2), Saliva collector for vial 2, Plastic vial containing B2 buffer (2 components): specifications yet to be added.
Oligonucleotide S1: 5’- CGGTAGTAGCCAAGCACCCATGTTAGACCTC - 3’
Oligonucleotide S2: 5’ – CCTGCTCAGCGATCTATTTGGTCATCTGG -3’
Oligonucleotide H1: 5’– GGGTAGGGCGGGTTGGGAGAGGTCAGAGGTCTr(A)GAGCAGGGT TACACCCATGTGACCTCTCCC – 3’
Oligonucleotide H2: 5’ – ACCCCCTGCTCAGCGATTAACGAGGTCTr(A)GAGCAGGAGCAG GGGGTAGGGCGGGTTGGG A - 3’
Oligonucleotide Linker: 5’ – GACCTCCCTGCT – 3’
Oligonucleotide FreeHRP: 5' - GGGTAGGGCGGGTTGGGAGAGGTCAGAGGTCT - 3'
Upon arrival all oligonucleotides are redissolved with a 20 mM pH 7.4 Tris-HCl containing 1 mM EDTA. Prior to use, the H1 and H2 solutions were first heated for 5 min at 95 C followed by cooling to room temperature with a ramp of 1°C per minute. The S1 and S2 solutions were first heated for 5 min at 95 C then cooled on ice.
Buffer B1: 100 mM NaCl, 10 mM MgCl2, 10 mM KCl, 100 nM H1, 100 nM H2, 50 nM S1, 50 nM S2, 60 nM Linker (according to Xie et al., 2020), 10mM TrisHCl pH 7.5 Tris, proteinase K to 0.05 mg/ml and SDS to 10%. Supplied dryed.
Buffer B2: 80 mg/l of tetrachloroauric(III) acid (HAuCl4) in 1mM MES. Supplied as 2 components to be mixed.
Test box: a polystyrene printed as in fig.1. It includes one loading pad, three substrate pads and three reagent pads. The loading pad is a 50 mm cellulose disk. The substrate pads are similar but have been soaked in a 5 mM glucose solution then dried before mounting on the box. The reagent pads are similar but receive a different pretreatment before being mounted as T (test), C+ (positive control) and C- (negative control) as in fig.1: C+ is soaked in a 0.1 nM solution of FreeHRP oligo, dried, soaked in a hemin solution (25 µM in 25 mM HEPES pH 7.4 containing 100 mM NaCl, 10 mM MgCl2, 10 mM KCl, 0.05% Triton X-10 and 1% DMSO), dried, soaked in a solution of glucose and then dried again; T is not soaked in the oligo; C- is not soaked in glucose. The bottom of the box is a removable sheet of Whatman #1 cellulose paper, photolitographed according to fig.2. After removal of the paper layer the box is sealed to define the reaction wells.
Arunachalam S. R., Tang K. D., Punyadeera C. (2019). Isolation and Quantification of MicroRNAs from Human Saliva. InTheranostics(pp. 105-114). Humana, New York, NY.
Calabria D., Caliceti C., Zangheri M., Mirasoli M., Simoni P., Roda A. (2017). Smartphone-based enzymatic biosensor for oral fluid L-lactate detection in one minute using confined multilayer paper reflectometry. Biosens Bioelectron. 94:124-130.
Chen Z., Xiong F., Yu A., Lai G. (2019). Aptamer biorecognition-triggered DNAzyme liberation and Exo III-assisted target recycling for ultrasensitive homogeneous colorimetric bioassay of kanamycin antibiotic. Chemical communications, 55(27), 3959-3962.
Chu D. K., Pan Y., Cheng S., Hui K. P., Krishnan P., Liu Y., Ng D., Wan C., Yang P., Wang Q., Peiris M., Poon L. (2020). Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia.Clinical chemistry.
de la Rica R., Stevens, MM. (2012) Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nature Nanotech 7, 821-824.
de la Rica R, Stevens MM. (2013) Plasmonic ELISA for the detection of analytes at ultralow concentrations with the naked eye. Nat Protoc. 8:1759-64.
Fronczek A., Park T.S., Harshman D.K., Nicolinia A.M., Yoon J.-Y. (2014). Paper microfluidic extraction and direct smartphone-based identification of pathogenic nucleic acids from field and clinical samples. RSC Adv. 4: 11103.
Li W, Li Y, Liu Z, Lin B, Yi H, Xu F, Nie Z, Yao S. (2016) Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res. 44:7373-84.
Low S. S., Pan Y., Ji D., Li Y., Lu Y., He Y., Chen Q., Liu Q. (2020). Smartphone-based Portable Electrochemical Biosensing System for Detection of Circulating MicroRNA-21 in Saliva as a Proof-of-Concept.Sensors and Actuators B: Chemical, 127718.
Martinez, A. W.; Phillips, S. T.; Butte, M. J.; Whitesides, G. M. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318-1320.
Mokany E, Bone SM, Young PE, Doan TB, Todd AV. (2010) MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc. 132:1051-9.
Pfaffe T., Cooper-White J., Beyerlein P., Kostner K., Punyadeera, C. (2011). Diagnostic potential of saliva: current state and future applications.Clinical chemistry,57(5), 675-687.
Safdar S., Ven K., van Lent J., Pavie B., Rutten I., Dillen A., Munck S., Lammertyn J., Spasic D. (2020). DNA-only, microwell-based bioassay for multiplex nucleic acid detection with single base-pair resolution using MNAzymes.Biosensors and Bioelectronics, 112017.
Soda Y., Bakker E. (2019) Quantification of Colorimetric Data for Paper-Based Analytical Devices. ACS Sens 4:3093−3101
To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A. R., Wu T.-C., et al. (2020a). Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis https://doi.org/10.1016/S1473-3099(20)30196-1.
To K.K.-W., Tsang O.T.-Y., Yip C. C.-Y., Chan, K.-H., Wu T.-C., et al. (2020b) Consistent Detection of 2019 Novel Coronavirus in Saliva, Clinical Infectious Diseases, ciaa149, https://doi.org/10.1093/cid/ciaa149
Ven K, Safdar S, Dillen A, Lammertyn J, Spasic D. Re-engineering 10-23 core DNA- and MNAzymes for applications at standard room temperature. Anal Bioanal Chem. 2019 Jan;411(1):205-215. doi: 10.1007/s00216-018-1429-4
Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. Published online March 11, 2020. doi:10.1001/jama.2020.3786
Xie Y., Niu F., Yu A., Lai G. (2020). Proximity Binding-Triggered Assembly of Two MNAzymes for Catalyzed Release of G‑Quadruplex DNAzymes and an Ultrasensitive Homogeneous Bioassay of Platelet-Derived Growth Factor. Anal. Chem., 92, 1, 593-598
The report on lab testing work on prototype v0.1b is available for download
Giuseppe Firrao, Sabrina Palmano, Giulia Tarquini, Federico Bosetto, Rita Musetti, Gaia Carminati, Francesco Savian