Release version 0.2a
Summary description
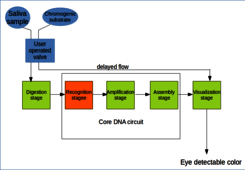
The biosensor will be a simple, single use plastic microfluidic device that will be loaded with a drop of saliva by the user and then will perform mixing with reaction components with simple gestures of the user. Upon completion of the reaction time the device will put the reaction results into contact with the chromogenic solution. The reaction is being carried out almost entirely from sampling to results in the same solution with no need to washes, buffer changes, purification or any other action requiring supervision, despite the presence of different stages, i.e. consecutive processing steps that build up a eye detectable signal from the input sample.
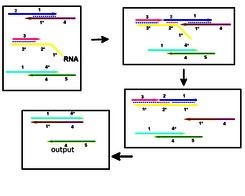
The biosensor core is a DNA circuit that will include three stages. The first stage will be a cooperative strand displacement reaction that is specifically initiated by the viral RNA. This “recognition” module will output a single stranded DNA fragment with a DNA oligonucleotide with standard (S1) sequence that will be used by the following module as input. The second stage will be a well assessed, robust, low background amplification module. Since the input of this stage is a standard sequence, it will not be necessary to adapt or newly develop an “amplification” process: conversely a well established, widely tested, robust method will be adopted. The initiator sequence that proved to work best with the amplifier will be set to be S1, as the output of the “recognition” stage can be chosen without constrain. The amplification stage will output a DNA oligonucleotide (S2) that will act as template for the hybridization of two halves of a split G-quadruplex HRP-mimiking DNAzyme, to be thus assembled in a functional molecule with peroxidation properties (“assembly” stage). Also in this case, there will be no targeted project and the results of previously published research in the field will be exploited, as the best way to split DNAzymes has been deeply investigated.
The biosensor core will be completed with a preliminary “digestion” stage and a final “visualization” stage. In the former, the nucleic acid target, i.e. the SARS-CoV2 RNA will be released in the solution and made available for the further steps by the action of lysis agents such as SDS and proteinase K. In the “visualization” stage a chromogenic substrate solution is added to the reaction, allowing the peroxidase activity to develop a eye detectable color.